This chapter explains how to use performance tools to reduce the execution time of your programs. This chapter describes prof, pixie, and cord. For information about the compiler optimization options, see Chapter 5, "Optimizing Program Performance."
This chapter covers the following topics:
"Overview of Profiling" explains how profiling can help you to analyze your data.
"Profiling With prof" describes how to run the profiler, prof and lists its options.
"pc Sampling" explains how to use prof to obtain program counter (pc) sampling.
"Basic Block Counting" covers how to use prof and pixie perform basic block counting.
"Profiling Multiprocessed Executables" describes how to profile executables that use sproc and sprocsp system calls.
"Rearranging Procedures With cord" explains how to rearrange procedures to reduce paging and achieve better instruction cache mapping.
Although it may be possible to obtain short-term speed increases by relying on unsupported or undocumented quirks of the compiler system, it's a bad idea to do so. Any such "features" may break in future releases of the compiler system.
The best way to produce efficient code that can be trusted to remain efficient is to follow good programming practices; in particular, choose good algorithms and leave the details to the compiler.
The techniques described in this manual comprise only a part of performance tuning. Other areas that you can tune, but are outside the scope of this document, include graphics, I/O, the kernel, system parameters, memory, and real-time system calls.
Profiling is a three-step process that consists of compiling the source program, executing the program, and then running the profiler, prof, to analyze the data.
The compiler system provides two types of profiling:
Program counter (pc) sampling, which measures the amount of execution time spent in various parts of the program. This statistical data is obtained by periodically sampling the program counter. For example, cc –p interrupts the program ever 10 milliseconds, and records the value of the program counter. By default, prof generates pc sampling data.
Basic block counting, which counts the execution of basic blocks (a basic block is a sequence of instructions that is entered only at the beginning and exits only at the end). It produces an exact count of the number of times each basic block is executed, thereby providing more detailed information than pc sampling.
Profiling produces detailed information about program execution. You can use profiling tools to find the areas of code where most of the execution time is spent. In a typical program, a large part of the execution time is spent in relatively few sections of code. It is a good idea to concentrate on improving code efficiency in those sections first.
The topics covered below include:
The profiler program, prof(1), analyzes raw profiling information and produces a printed report. The program analyzes either pc sampling or basic block counting data.
The syntax for prof is:
prof [options] [prog_name] [profile_filename ...] |
| options | One of the keywords or keyword abbreviations shown in Table 4-1. (Specify either the entire name or the initial character of the option, as indicated in the table.) | |
| prog_name | Specifies the name of the program whose profile data is to be profiled. | |
| profile_filename | Specifies one or more files containing the profile data gathered when the profiled program executed (defaults are explained below). If you specify more than one file, prof sums the statistics in the resulting profile listings. |
The prof program has these defaults:
If you do not specify –pixie, prof assumes pc-sampling data is being analyzed. If you do not specify profile_filename, the profiler looks for a mon.out file. If this file does not exist in the current directory, prof looks for profile input data files in the directory specified by the PROFDIR environment variable (see "Creating Multiple Profile Data Files" for information on PROFDIR).
You may want to use the –merge option when you have more than one profile data file. This option merges the data from several profile files into one file.If you specify –pixie and do not specify profile_filename, then prof looks for prog_name.Counts and provides basic block count information if this file is present.
If you specify profile_filename(s), prof determines the file type based on its content: a prof- or pixie-mode file.
Table 4-1 lists prof options. Options that apply to basic block counting are indicated as such.
Program counter (pc) sampling reveals the amount of execution time spent in various parts of a program. The count includes:
CPU time and memory access time
Time spent in user routines
The pc sampling does not count time spent swapping or time spent accessing external resources.
This section explains how to obtain pc sampling and provides examples showing the use of various prof options. Specifically, this section covers:
Obtain pc sampling information by linking the desired source modules using the –p option and then executing the resulting program object, which generates raw profile data.
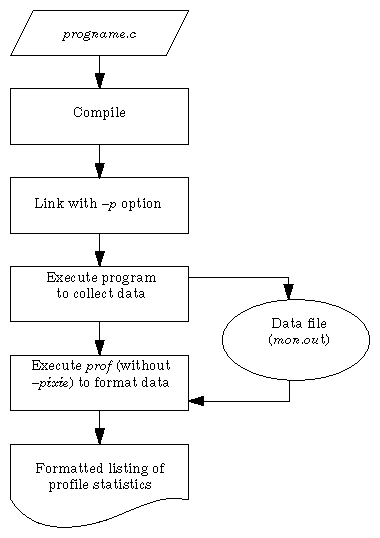
Use the procedure below to obtain pc sampling information. Also refer to Figure 4-1, which illustrates how pc sampling works.
Compile the program using the appropriate compiler. For example, to compile a C program myprog.c:
% cc -c myprog.c
Link the object file created in Step 1.
% cc -p -o myprog myprog.o

Note: You must specify the –p profiling option during this step to obtain pc sampling information. Execute the profiled program (just as you would execute an unprofiled program).
% myprog
During execution, profiling data is saved in the file mon.out. You can run the program several times, altering the input data, to create multiple profile data files. You can also use the environment variable PROFDIR as explained in "Creating Multiple Profile Data Files."
Run the profile formatting program prof.
% prof -pcsample myprog mon.out
prof extracts information from mon.out and prints it in an easily readable format. If mon.out exists, it is overwritten. Therefore, rename each mon.out to save its output. For more information, see the prof(1) reference page.
Include or exclude information on specific procedures within your program by using the –only or –exclude profiler options (refer to Table 4-1).
When you run a program using pc sampling, raw data is collected and saved in the profile data file mon.out. To collect profile data in several files, or to specify a different name for the profile data file, set the environment variable PROFDIR, using the appropriate method from Table 4-2.
Table 4-2. Setting a PROFDIR Environment Variable
C shell and tcsh | Bourne shell and Korn shell |
|---|---|
setenv PROFDIR dirname | PROFDIR=dirname; export PROFDIR |
Setting the environment variable puts the raw profile data of each invocation of progname in files named dirname/progname.mon.pid. (You must create a directory named dirname before you run the program.) pid is the process ID of the executing program; progname is the name of the program when invoked.
The default frequency of pc sampling is 10 milliseconds. You can change the pc sampling to 1 millisecond by setting the environment variable:
PROF_SAMPLING=1 |
Since pc sampling is statistical, this provides more accurate profiling data. However, be aware that considerable kernel overhead is incurred for every process executing on the system while the profiled program is running.
The examples in this section illustrate how to use prof and its options to obtain pc sampling data.
The following partial listing is an example of pc sampling output from a profiled version of the program test.
--------------------------------------------------------------------------
Profile listing generated Tue Oct 4 14:16:30 1994
with: prof -pcsample test test.mon.207
--------------------------------------------------------------------------
samples time CPU FPU Clock N-cpu S-interval Countsize
7473 75s R4000 R4010 100.0MHz 0 10.0ms 0(bytes)
Each sample covers 4 bytes for every 10.0ms ( 0.01% of 74.7300sec)
--------------------------------------------------------------------------
-p[rocedures] using pc-sampling.
Sorted in descending order by the number of samples in each procedure.
Unexecuted procedures are excluded.
--------------------------------------------------------------------------
samples time(%) cum time(%) procedure (file)
3176 32s( 42.5) 32s( 42.5) _cerror (/usr/lib/libc.so.1:cerror.s)
2564 26s( 34.3) 57s( 76.8) _doprnt (/usr/lib/libc.so.1:doprnt.c)
578 5.8s( 7.7) 63s( 84.5) _isatty (/usr/lib/libc.so.1:isatty.c)
441 4.4s( 5.9) 68s( 90.4) offtime (/usr/lib/libc.so.1:time_comm.c)
217 2.2s( 2.9) 70s( 93.3) atoi (/usr/lib/libc.so.1:atoi.c)
... |
In the above listing:
The samples column reports the number of samples in each procedure, sorted in descending order. For example, there were 3176 samples for the procedure _cerror.
The time(%) column lists the number of seconds and percentage of execution time spent in each procedure. For example, there were 32 seconds (42.5% of execution time) spent in _cerror.
The cum time(%) column lists the percentage of the total execution time spent in each procedure. For example, there were 63 seconds (84.5% of total execution time) were spent cumulatively in the _cerror, _doprnt, and _isatty procedures. Note that this does not imply that these routines called each other; they may have executed sequentially.
The procedure(file) column prints the procedure name and its source file. For example, the source file containing the _cerror procedure is /usr/lib/libc.so.1:cerror.s.
You can use the –dis option to prof to disassemble the analyzed object code and see the number of cycles it takes to execute an instruction. Check the disassembled code for stalls (wasted cycles) and the number of instructions per cycle. For example, partial output looks like this:
Profile listing generated Tue Oct 4 13:43:41 1994
with: prof -pixie -dis hello
-----------------------------------------------------------------------------
Total cycles Total Time Instructions Cycles/inst Clock Target
5005674 0.05006s 3003856 1.666 100.0MHz R4000
28 cycles due to code that could not be
assigned to any source procedure.
1000547: Total number of Load Instructions executed.
4001532: Total number of bytes loaded by the program.
200261: Total number of Store Instructions executed.
800774: Total number of bytes stored by the program.
100223: Total number nops executed in branch delay slot.
200558: Total number conditional branches executed.
200418: Total number conditional branches actually taken.
0: Total number conditional branch likely executed.
0: Total number conditional branch likely actually taken.
601965: Total cycles waiting for current instr to finish.
4202824: Total cycles lost to satisfy scheduling constraints.
3001119: Total cycles lost waiting for operands be available.
---------------------------------------------------------------------------
* -p[rocedures] using basic-block counts. *
* Sorted in descending order by the number of cycles executed in each *
* procedure. Unexecuted procedures are not listed. *
----------------------------------------------------------------------------
cycles(%) cum % secs instrns calls procedure(file)
5000044(99.89) 99.89 0.05 3000032 1 main(hello:hello.c)
1566( 0.03) 99.92 0.00 985 3 fflush(/usr/lib/libc.so.1:flush.c)
1332( 0.03) 99.95 0.00 861 1 _doprnt(/usr/lib/libc.so.1:doprnt.c)
1130( 0.02) 99.97 0.00 1004 2 _dtoa(/usr/lib/libc.so.1:dtoa.s)
320( 0.01) 99.97 0.00 120 4_dwmultu(/usr/lib/libc.so.1:tenscale.s)
302( 0.01) 99.98 0.00 196 4 memcpy(/usr/lib/libc.so.1:bcopy.s)
...
------------------------------------------------------------------------------
* -dis[assemble] listing annotated with cycle counts. *
* Unexecuted procedures are excluded. *
------------------------------------------------------------------------------
crt1text.s
__start: <0x400900-0x400a08>
77 total cycles(0.00%) invoked 1 times, average 77 cycles/invocation
[91] 0x00400900 0x03e04025 or t0,ra,0 # 1
[91] 0x00400904 0x04110001 bgezal zero,0x40090c # 2
[91] 0x00400908 0000000000 nop # 3
<2 cycle stall for following instruction>
^--- 5 total cycles(0.00%) executed 1 times, average 5 cycles.---^
[91] 0x0040090c 0x3c1c0fc0 lui gp,0xfc0 # 6
... |
The previous listing shows statistics about the file hello. The statistics detail procedures using basic-block counts and disassembled code. Information at the top of the listing is self-explanatory. Of interest are cycles waiting and cycles lost.
The –p[rocedures] information uses basic-block counts to sort in descending order the number of cycles executed in each procedure.
The cycles(%) column lists the number of cycles (and percentage of total cycles) per procedure. For example, there were 5000044 cycles (or 99.89%) for the procedure main.
The cum% column shows the cumulative percentage of cycles. For example, main used 99.89% of all cycles.
The secs column reports the number of seconds spent in the procedure. For example, 0.05 seconds were spent in main.
The instrns column lists the number of instructions executed in the procedure. For example, 3000032 instructions were executed in main.
The calls procedure(file) column shows the number of calls in the procedure. For example, there was 1 call in main.
The –dis[assemble] information provides a listing containing cycle counts. It lists the beginning and ending addresses of crt1text.s __start: <0x400900-0x400a08>. It also reports the total cycles for a procedure, number of times invoked, and average number of cycles per invocation:
77 total cycles(0.00%) invoked 1 times, average 77 cycles/invocation
The first column lists the line number of the instruction: [91]
The second column lists the beginning address of the instruction: 0x00400900
The third column shows the instruction in hexadecimal: 0x03e04025.
The fourth column reports the assembler form (mnemonic) of the instruction: or t0,ra,0
The last column reports the cycle in which the instruction executed: #1
Other information includes:
The total number of cycles in a basic block and the percentage of the total cycles for that basic block, the number of times the branch terminating that basic block was executed, and the number of cycles for one execution of that basic block:
5 total cycles(0.00%) executed 1 times, average 5 cycles.Any cycle stalls (cycles that were wasted):
<2 cycle stall for following instruction>
For information on cycle stalls and what causes them, see the MIPS Microprocessor Chip Set User's Guide for your architecture.
Basic block counting, obtained using the program pixie, measures the execution of basic blocks. A basic block is a sequence of instructions that is entered only at the beginning and exits only at the end. This section covers:
Use pixie(1) to measure the frequency of code execution. pixie reads an executable program, partitions it into basic blocks, and writes (instruments) an equivalent program containing additional code that counts the execution of each basic block.
Note that the execution time of an instrumented program is two-to-five times longer than an uninstrumented one. This timing change may alter the behavior of a program that deals with a graphical user interface (GUI), or depends on events such as SIGALARM that are based on an external clock.
The syntax for pixie is:
pixie prog_name [options] |
| prog_name | Name of the input program. | |
| options | One of the keywords listed in Table 4-3. |
Table 4-3 lists pixie options. For a complete list of options refer to the pixie(1) reference page.
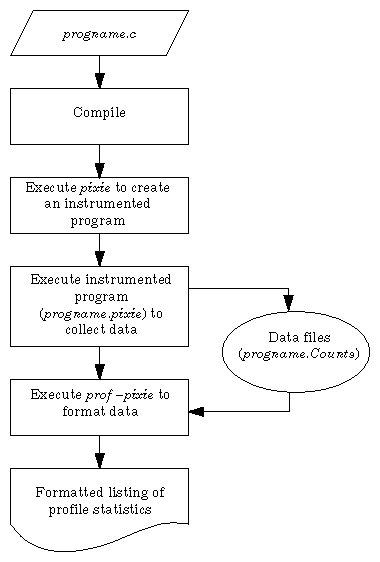
Use this procedure to obtain basic block counts. Also refer to Figure 4-2, which illustrates how basic block counting works.
Compile and link your program. Do not use the –p option. The following example uses the input file myprog.c.
% cc -o myprog myprog.c
The cc compiler compiles myprog.c into an executable called myprog.
Run pixie to generate the equivalent program containing basic-block-counting code.
% pixie myprog
pixie takes myprog and writes an equivalent program, myprog.pixie, containing additional code that counts the execution of each basic block. pixie also writes an equivalent program for each shared object used by the program (in the form: libname.so.pixie), containing additional code that counts the execution of each basic block. For example, if myprog uses libc.so.1, pixie generates libc.so.1.pixie.
Set the path for your .pixie files. pixie uses the rld search path for libraries (see rld(1) for the default paths). If the .pixie files are in your local directory, set the path as:
% setenv LD_LIBRARY_PATH .
Execute the file(s) generated by pixie (myprog.pixie) in the same way you executed the original program.
% myprog.pixie
This program generates a list of basic block counts in files named myprog.Counts. If the program executes a fork/sproc, a process ID is appended to the end of the filename (for example, myprog.Counts.345) for each process.
Run the profile formatting program prof specifying the –pixie option and the name of the original program.
% prof -pixie myprog myprog.Counts
prof extracts information from myprog.Counts and prints it in an easily readable format. If multiple .Counts files exist, you can use the wildcard character (*) to specify all of the files.
% prof -pixie myprog myprog.Counts*
 | Note: Specifying myprog.Counts is optional; prof searches by default for names having the form prog_name.Counts. |
You can run the program several times, altering the input data, to create multiple profile data files. See "Example Using prof –pixie –procedures –clock" later in this section for an example.
The time computation assumes a "best case" execution; actual execution may take longer. This is because the time includes predicted stalls within a basic block, but not actual stalls that may occur entering a basic block. Also it assumes that all instructions and data are in cache (for example, it excludes the delays due to cache misses and memory fetches and stores).
The complete output of the –pixie option is often extremely large. Use the –quit option with prof to restrict the size of the output. Refer to "Running the Profiler" for details about prof options.
Include or exclude information on specific procedures in your program by using the prof options –only or –exclude (explained in Table 4-1). prof timings reflect only time spent in a specific procedure, not time spent including procedures called by that procedure. The CASEVision/WorkShop toolset, an optional software product, can show an estimate of inclusive times.
The examples in this section illustrate how to use prof –pixie to obtain basic block counting information from a profiled version of a C file, espresso.
The partial listing below illustrates the use of the –i[ nvocations] option. For each procedure, prof reports the number of times it was invoked from each of its possible callers and lists the procedure(s) that called it.
% prof -pixie -i espresso |
-------------------------------------------------------------------------------
Profile listing generated Fri May 13 14:25:19 1994
with: prof -pixie -i espresso
...
* Sorted in descending order by number of calls per procedure.
* Unexecuted procedures are excluded.
* The inst column is a static number of instructions.
* %coverage column contains the percent inst executed.
-------------------------------------------------------------------------------
Total procedure invocations: 12113082
calls(%) cum% inst %coverage procedure (file)
3055229(25.22) 25.22 26 25.00 full_row
(espresso:/usr/people/guest/enjoy/008.espresso/setc.c)
966541( 7.98) 33.20 26 25.00 set_or
(espresso:/usr/people/guest/enjoy/008.espresso/set.c)
772942( 6.38) 39.58 26 25.00 cleanfree
(espresso:/work/irix/lib/libc/gen/malloc.c)
611793( 5.05) 44.63 26 25.00 setp_implies
... |
The above listing shows the total procedure invocations (calls) during the run: 12113082.
The calls(%) column reports the number of calls (and the percentage of total calls) per procedure. For example, there were 3055229 calls (or 25.22% of the total) spent in the procedure full_row.
The cum% column shows the cumulative percentage of calls. For example, 25.22% of all calls were spent in full_row.
The inst column shows the number of instructions executed for a procedure. For example, there were 26 instructions in the procedure full_row.
The %coverage column reports the percentage of instructions executed. For example, 25.00% of instructions were executed in full_row.
The procedure (file) column lists the procedure and its file. For example, the first line reports statistics for the procedure full_row in the file setc.c.
The following partial listing shows the source code lines responsible for the largest portion of execution time produced with the –heavy option.
% prof -pixie -heavy espresso |
-------------------------------------------------------------------------------
Profile listing generated Fri May 13 14:28:56 1994
with: prof -pixie -heavy espresso
-------------------------------------------------------------------------------
...
* -h[eavy] using basic block counts.
* Sorted in descending order by number of cycles per line.
* Unexecuted lines are excluded.
* The insts column contains distinct executed instructions for this line.
-------------------------------------------------------------------------------
cycles(%) cum % line insts procedure (file)
77948528( 4.95%) 4.95% 57 40 cofactor
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
73800963( 4.69%) 9.65% 213 67 essen_parts
(espresso:/usr/people/guest/enjoy/008.espresso/expand.c)
53399667( 3.39%) 13.04% 48 29 full_row
(espresso:/usr/people/guest/enjoy/008.espresso/setc.c)
44723520( 2.84%) 15.88% 137 22 massive_count
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
38032848( 2.42%) 18.30% 257 39 taut_special_cases
(espresso:/usr/people/guest/enjoy/008.espresso/irred.c)
... |
The previous partial listing shows basic block counts sorted in descending order. The most heavily used line (57) was in procedure cofactor.
The cycles(%) column shows the total number of program cycles (and percentage of the total number). For example, there were 77948528 cycles (4.95% of the total number) for the procedure cofactor.
The cum% column shows the cumulative percentage of the total program cycles. For example, 4.95% of all program cycles were spent in cofactor. The first three procedures used 13.04% of the total cycles.
The line column lists the line number of the procedure: 57.
The insts column reports the number of distinct instructions that were executed at least once. For example, line 57 had 40 instructions.
The following partial listing, produced using the –lines option, shows the execution time spent on each line of code, grouped by procedure.
% prof -pixie -lines espresso |
-------------------------------------------------------------------------------
Profile listing generated Fri May 13 14:30:49 1994
with: prof -pixie -lines espresso
-------------------------------------------------------------------------------
...
* -l[ines] using basic-block counts.
* Grouped by procedure.
* Major sort on cycles executed per procedure.
* Minor sort on line numbers within procedure.
* Unexecuted procedures are execluded.
-------------------------------------------------------------------------------
cycles(%) cum % line insts procedure (file)
856768( 0.05%) 0.05% 121 12 massive_count
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
25235712( 1.60%) 1.66% 128 12 massive_count
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
934656( 0.06%) 1.72% 134 16 massive_count
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
5963136( 0.38%) 2.10% 135 7 massive_count
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
20870976( 1.33%) 3.42% 136 13 massive_count
(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
... |
In the above listing:
The cycles(%) column lists the number of program cycles (and the percentage of the total cycles) for each procedure. For example, there were 856768 program cycles (0.05% of the total) for massive_count.
The cum% column shows the cumulative percentage of the total program cycles. For example, 0.05% of all program cycles were spent in massive_count.
The line and insts columns report the procedure's line number, and number of distinct instructions. For example, for the procedure massive_count, lines 121 and 128 each generated 12 instructions that were executed at least once, and line 134 generated 16 instructions that were executed at least once.
You can limit the output of prof to information on only the most time-consuming parts of the program by specifying the –quit option. You can instruct prof to quit after a particular number of lines of output, after listing the elements consuming more than a certain percentage of the total, or after the portion of each listing whose cumulative use is a certain amount.
Consider the following sample listing:
cycles(%) cum % seconds cycles bytes procedure(file)
/call /line
360331656(22.90) 22.90 4.80 4626 93
massive_count(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
174376925(11.08) 33.99 2.33 15479 108
cofactor(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
157700711(10.02) 44.01 2.10 43817 123
elim_lowering(espresso:/usr/people/guest/enjoy/008.espresso/expand.c)
155670642( 9.89) 53.91 2.08 49216 156
essen_parts(espresso:/usr/people/guest/enjoy/008.espresso/expand.c)
66835754( 4.25) 58.15 0.89 691 76
scofactor(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
66537017( 4.23) 62.38 0.89 21 156
full_row(espresso:/usr/people/guest/enjoy/008.espresso/setc.c)
57747597( 3.67) 66.05 0.77 1670 87
taut_special_cases(espresso:/usr/people/guest/enjoy/008.espresso/irred.c) |
Any one of the following commands eliminates everything from the line starting with 66835754 to the end of the listing:
prof -quit 4 prof -quit 5% prof -quit 53cum% |
The following partial listing, produced with the –procedures option, shows the percentage of execution time spent in each procedure.
% prof -pixie -procedures espresso |
-------------------------------------------------------------------------------
Profile listing generated Fri May 13 14:33:00 1994
with: prof -pixie -procedures espresso
-------------------------------------------------------------------------------
...
* -p[rocedures] using basic-block counts.
* Sorted in descending order by the number of cycles executed in each
* procedure. Unexecuted procedures are not listed.
-------------------------------------------------------------------------------
cycles(%) cum % seconds cycles bytes procedure(file)
/call /line
360331656(22.90) 22.90 4.80 4626 93
massive_count(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
174376925(11.08) 33.99 2.33 15479 108
cofactor(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
157700711(10.02) 44.01 2.10 43817 123
elim_lowering(espresso:/usr/people/guest/enjoy/008.espresso/expand.c)
155670642( 9.89) 53.91 2.08 49216 156
essen_parts(espresso:/usr/people/guest/enjoy/008.espresso/expand.c)
66835754( 4.25) 58.15 0.89 691 76
scofactor(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c) |
In the above listing:
The cycles(%) column lists the number of program cycles (and percentage of the total) used. For example, massive_count used 360331656 program cycles (22.90% of the total cycles).
The cum% column reports the cumulative total of all cycles used. For example, massive_count, cofactor, and elim_lowering used 44.01% of the cycles.
The seconds column lists the time used by the procedure (the clock rate, 75 megahertz, is omitted in this example). For example, there were 4.80 seconds used by massive_count.
The cycles/call and bytes/line columns report the average cycles per call, and the bytes of code per line of source text, respectively. For example, massive_count used an average of 4626 cycles per call, and had 93 bytes of generated code per line of source text.
The procedure (file) column lists the procedure, massive_count, and its source file, cofactor.c.
You can add absolute time information to the output by specifying the clock rate, in megahertz, with the –clock option. Partial output looks like this:
% prof -pixie -procedures -clock 20 espresso |
-------------------------------------------------------------------------------
Profile listing generated Fri May 13 14:34:55 1994
with: prof -pixie -procedures -clock 20 espresso
-------------------------------------------------------------------------------
...
* -p[rocedures] using basic-block counts.
* Sorted in descending order by the number of cycles executed in each
* procedure. Unexecuted procedures are not listed.
-------------------------------------------------------------------------------
cycles(%) cum % seconds cycles bytes procedure(file)
/call /line
360331656(22.90) 22.90 18.02 4626 93
massive_count(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
174376925(11.08) 33.99 8.72 15479 108
cofactor(espresso:/usr/people/guest/enjoy/008.espresso/cofactor.c)
157700711(10.02) 44.01 7.89 43817 123
elim_lowering(espresso:/usr/people/guest/enjoy/008.espresso/expand.c)
... |
In the previous listing, the seconds column lists the number of seconds spent in each procedure. For example 18.02 seconds were spent in the procedure massive_count. The time (computed by the profiler), in seconds, is based on the machine speed specified with the –clock option (in megahertz). In this example, the speed specified is 20 megahertz.
Sometimes a single run of a program does not produce the results you require. You can repeatedly run the version of a program created by pixie and vary the input with each run, then use the resulting .Counts files to produce a consolidated report.
Use the following procedure to average prof results:
Compile and link the input file. Do not use the –p option. For example, consider the input file myprog.c:
% cc -o myprog myprog.c
The cc compiler compiles myprog.c and saves the executable as myprog.
Run the profiling program pixie.
% pixie myprog -pids
pixie generates the modified program myprog.pixie.
Run the profiled program as many times as desired. Each time you run the program, pixie creates a myprog.Counts.pid file, with the process ID appended.
% myprog.pixie < input1 > output1 % myprog.pixie < input2 > output2 % myprog.pixie < input3 > output3
Create the report.
% prof -pixie myprog myprog.Counts*
prof sums the basic block data in the .Counts files to produce the profile report.
Use pixstats(1) to get more architectural details of a program's execution than are available from prof. The –op option to pixstats produces low-level information on bus issue, various kinds of stalls that prof doesn't provide. Prof also requires more memory to operate, so in situations where not enough memory exists for prof to function correctly, you can use pixstats.
You can also use pixstats to look for write buffer stalls and to produce disassembled code listings.
 | Note: In subsequent releases, pixstats will be removed and its functionality will be moved into prof. |
The disadvantages to using pixstats are that it:
Does not provide a line-by-line count
Profiles only one .Counts file at a time (no averaging)
Provides very little documentation
Does not show time spent in floating point exceptions
The syntax for pixstats is:
pixstats program [options] |
| program | Specifies the name of the program to be analyzed. | |
| options | One of the keywords shown in Table 4-4. |
Table 4-4 lists and briefly describes pixstats options. For details, see the pixstats(1) reference page.
Table 4-4. Options for pixstats
Name | Result |
|---|---|
–cycle ns | Assumes an ns cycle time when converting cycle counts to seconds. |
–dis | Disassembles and annotates the analyzed object code. |
–dso [dso_name] | Analyzes only the named DSO(s). |
–onlyprocedure_name | Analyzes only the named procedure(s). |
–op | Produces a detailed listing about instructions and operations and procedure usage. Information includes instruction distribution, stall distribution, basic block size distribution, and register usage. |
–r2010 | Uses r2010 floating point chip operation times and overlap rules. This option is the default. |
–r2360 | Uses r2360 floating point board operation times and overlap rules. |
–r4000 | Uses the r4000 operation times and overlap rules. This is the default if the program's magic number indicates it is a mips2 or mips3 executable. |
Other options are explained in the pixstats(1) reference page.
Use the following procedure to run pixstats:
Compile and link the input file myprog.c. Do not use the –p option. For example, using the input file myprog.c:
% cc -c myprog.c % cc -o myprog myprog.o
The cc compiler driver compiles myprog.c and saves the object file as myprog.o. The second command links myprog.o and saves the executable as myprog.
Run the profiling program pixie.
% pixie myprog
pixie generates the modified program myprog.pixie.
Set the path, so pixie knows where to find the .pixie files.
% setenv LD_LIBRARY_PATH .
Execute the file generated by pixie, myprog.pixie, in the same way you would execute the original program.
% myprog.pixie
This program generates the file myprog.Counts which contains the basic block counts.
Run pixstats to generate a detailed report.
% pixstats myprog
The following example shows the default listing generated by pixstats:
pixstats espresso:
1588254395 (1.357/inst) cycles (15.88s @ 100.0MHz)
1170355761 (1.000/inst) instructions
2397 (0.000/inst) floating point ops (0.000151 MFLOPS @ 100.0MHz)
Procedures ordered by execution time:
cycles %cycles cum% instrs cycles calls cycles procedure
/inst /call
382093989 24.1% 24.1% 278174631 1.4 77888 4906 massive_count
194452825 12.2% 36.3% 130750578 1.5 11265 17262 cofactor
146765915 9.2% 45.5% 104525532 1.4 3599 40780 elim_lowering
144704109 9.1% 54.7% 113501194 1.3 3163 45749 essen_parts
65043668 4.1% 58.7% 51198838 1.3 96713 673 scofactor
57256404 3.6% 62.4% 41920736 1.4 34564 1657 taut_special_cases
54258762 3.4% 65.8% 43594130 1.2 1632626 33 full_row
43947988 2.8% 68.5% 32692126 1.3 72095 610 sccc_special_cases
42611632 2.7% 71.2% 27971390 1.5 2370 17980 setup_BB_CC
35769668 2.3% 73.5% 26776245 1.3 528962 68 __malloc
29107500 1.8% 75.3% 24012582 1.2 333396 87 cdist01
28840766 1.8% 77.1% 23333068 1.2 235410 123 force_lower
27458158 1.7% 78.8% 21150937 1.3 447933 61 realfree
26682338 1.7% 80.5% 21303304 1.3 407124 66 cleanfree
21207623 1.3% 81.9% 16338599 1.3 528945 40 __free
19991078 1.3% 83.1% 13678106 1.5 526081 38 _malloc
19464960 1.2% 84.3% 13152000 1.5 526080 37 _free
17434271 1.1% 85.4% 15501189 1.1 485880 36 set_or
14574313 0.9% 86.4% 9466949 1.5 725 20103 expand1_gasp
13115606 0.8% 87.2% 9753942 1.3 336135 39 _smalloc
10646822 0.7% 87.9% 7928278 1.3 316901 34 setp_implies
9812911 0.6% 88.5% 7858695 1.2 50103 196 binate_split_select
9487312 0.6% 89.1% 7085744 1.3 908 10449 compl_lift
9342972 0.6% 89.7% 6531388 1.4 567 16478 cb_consensus
8825424 0.6% 90.2% 7330700 1.2 55166 160 consensus
8353958 0.5% 90.7% 7474594 1.1 219841 38 set_diff
7858360 0.5% 91.2% 6439566 1.2 32735 240cb_consensus_dist0
7670120 0.5% 91.7% 5122330 1.5 72095 106 sccc
7606139 0.5% 92.2% 6589200 1.2 66552 114 cactive
6833384 0.4% 92.6% 4754352 1.4 105270 65 t_delete
6561606 0.4% 93.0% 4923904 1.3 274750 24 set_clear
6065768 0.4% 93.4% 4007453 1.5 44414 137 sm_insert
5738712 0.4% 93.8% 4397726 1.3 95491 60 sf_addset
5477719 0.3% 94.1% 3639951 1.5 117592 47 t_splay
5162590 0.3% 94.5% 3351656 1.5 789 6543 essen_raising
5134827 0.3% 94.8% 3968016 1.3 1 5134827 rm_contain
4737968 0.3% 95.1% 4006370 1.2 34838 136 sccc_merge
4611671 0.3% 95.4% 3079225 1.5 2120 2175 intcpy
3868020 0.2% 95.6% 3438240 1.1 107445 36 set_and
3862752 0.2% 95.9% 3338484 1.2 66552 58 sccc_cube |
In the above listing, the first line shows the total cycles used and the second line shows the total instructions. The third line shows the number of floating point operations.
You can use pixstats –op to generate a detailed listing about instructions and operations. Information includes instruction distribution, stall distribution, basic block size distribution, and register usage.
The following example shows a partial listing generated by executing pixstats –op on the C file, espresso.
% pixstats -op espresso
1588254395 (1.357/inst) cycles (15.88s @ 100.0MHz)
1170355761 (1.000/inst) instructions
12892539 (0.011/inst) instructions annulled by untaken branch likely
250767706 (0.214/inst) cycles lost on non-sequential fetch
152077341 (0.130/inst) alu/shift/load result interlock cycles
689848 (0.001/inst) multiply interlock cycles (12 cycles)
692925 (0.001/inst) divide interlock cycles (75 cycles)
513369 (0.000/inst) variable shift extra issue cycles (2 total issue)
29852 (0.000/inst) floating point result interlock cycles
48 (0.000/inst) floating point compare interlock cycles
71323 (0.000/inst) interlock cycles due to basic block boundary
42595822 (0.036/inst) nops
570540263 (0.487/inst) alu (including logicals, shifts, traps)
185728252 (0.159/inst) logicals (including moves and li)
72264550 (0.062/inst) shifts
294103332 (0.251/inst) loads
70309957 (0.060/inst) stores
364413289 (0.311/inst) loads+stores
120292374 (0.103/inst) load followed by load
364415843 (0.311/inst) data bus use
77171 (0.000/inst) partial word references
117123216 (0.100/inst) sp+gp load/stores
2397 (0.000/inst) floating point ops (0.000151 MFLOPS @ 100.0MHz)
48 (0.000/inst) floating point compares
0 (0.000/inst) overlapped floating point cycles
168207521 (0.144/inst) conditional branches
35852721 (0.031/inst) branch likelies
32916066 (0.028/inst) load in branch delay slot
20834011 (0.018/inst) branch to branch
13630671 (0.012/inst) branch to branch taken
5836780 (0.005/inst) branch to branch untaken
41296177 (0.035/inst) branch delay filled with target-1 instruction
67419786 (0.058/inst) untaken conditional branches
100787735 (0.086/inst) taken conditional branches
17365744 (0.015/inst) taken conditional branches with bnop
7598403 (0.006/inst) untaken conditional branches with bnop
15749049 (0.013/inst) direction-predicted conditional branches with bnop
125383991 (0.107/inst) non-sequential fetches
236116938 (0.202/inst) basic blocks
8578595 (0.007/inst) calls
8594643 (0.007/inst) non-R31 JR
3511926 (0.003/inst) addui opportunities
0 (0.000/inst) fp multiply/add opportunities
2220801 (0.002/inst) skip |
You can use pixstats to disassemble and annotate the analyzed object code. The next example shows pixstats –dis[assemble]. The file, espresso, was executed on an R4000 CPU; results will differ on other CPUs.
% pixstats -dis espresso
0 43 404618 8fbc0020 lw gp,32(sp)
^---11265 total cycles. Executed 11265 times, avg 1 (0.00107% of inst.)---^
0 43 40461c 0040f825 move ra,v0
1 43 404620 afa20030 sw v0,48(sp)
2 43 404624 8f858574 lw a1,-31372(gp)
<< 2 cycle interlock >>
5 43 404628 8ca50000 lw a1,0(a1)
<< 2 cycle interlock >>
8 46 40462c 28a10021 slti at,a1,33
9 46 404630 10200003 beq at,zero,0x404640
10 46 404634 24a2ffff addiu v0,a1,-1
<< branch taken 11265 times (100%) >>
<< possible 2 cycles branch penalty, total 22530, avg 2 >>
^---146445 total cycles. Executed 11265 times, avg 13 (0.0075% of inst.)---^
0 46 404638 10000003 b 0x404648
1 46 40463c 24020002 li v0,2
^--- 0 total cycles. Executed 0 times, avg 4 (0% of inst.)---^
0 46 404640 00021143 sra v0,v0,5
1 46 404644 24420002 addiu v0,v0,2
^---22530 total cycles. Executed 11265 times, avg 2 (0.00214% of inst.)---^ |
The second line lists the total number of cycles for basic block 8fbc0020, (listed in the previous line). Line six shows a 2-cycle interlock because of a load of register a1 (referenced in line seven).
Line 12 shows that a branch was taken 11,265 times, and that branches were taken 100% of the time. A possible branch penalty exists for every branch. Line 13 shows an average of 2 penalties occurred for a total of 22,530 penalties (a large number because the branch was always taken).
You can gather either basic block and pc sampling profile data from executables that use the sproc and sprocsp system calls, such as those executables generated by POWER Fortran and POWER C. Prepare and run the job using the same method as for uniprocessed executables. For multiprocessed executables, each thread of execution writes its own separate profile data file. View these data files with prof like any other profile data files.
The only difference between multiprocessed and regular executables is the way in which the data files are named. When using pc sampling, the data files for multiprocessed executables are named process_id.prog_name. When using pixie, the data files are named prog_name.Counts.process_id. This naming convention avoids the potential conflict of multiple threads attempting to write simultaneously to the same file.
The cord(1) command rearranges procedures in an executable object to reduce paging and achieve better instruction cache mapping. This section describes cord and covers the following topics:
cord prog_name [reorder_file ...] |
Use cord to rearrange procedures in an executable to correspond with an ordering provided in a reorder_file. Typically, the ordering is arranged either to minimize paging and/or to maximize the instruction cache hit rate.
The reorder file is produced by the –feedback option to prof (for information on prof and the –feedback option, see Table 4-1, Options for prof, or the prof(1) reference page). The default reorder file is named prog.fb, if you do not specify reorder_file.
You can specify multiple reorder files on the command line; the first reorder file has the highest priority in rearranging the order. Thus you can improve paging in different program phases providing that multiple feedback files are generated by executing different phases of the program or by executing the program with distinct input data that cause different regions of the program to be executed.
Table 4-5 lists and describes cord options. For details, refer to the cord(1) reference page.
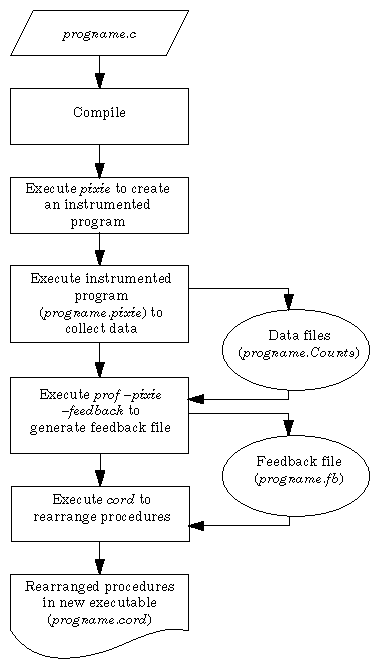
The example below shows how to use pixie, prof, and cord to rearrange the procedures in the program xlisp (refer to Figure 4-3).
% pixie xlisp # generates xlisp.pixie
% xlisp.pixie li-input.lsp # generates xlisp.Counts
% prof xlisp -pixie -feedback # generates xlisp.fb and
# libc.so.1.fb
% cord xlisp # generates xlisp.cord |
First, the program xlisp is executed by pixie, which generates an instrumented executable, xlisp.pixie. Next, the instrumented executable is run (with an input file to xlisp, li-input.lsp). Then prof is used to produce feedback files from the output data. Finally, cord is executed (and uses the order in the feedback file) to reorder the procedures in xlisp, generating a new binary, xlisp.cord. Figure 4-3 shows this procedure.
The procedure rearrangement depends on the data produced by the profiled runs of the executable. If these profiled runs approximate the actual use of the executable, the resultant binary is close to being optimally rearranged. Design your profiled runs accordingly.
You can also manually optimize your reorder file by rearranging the procedure entries in the reorder file.
For example, after running cord xlisp -pixie -feedback, the feedback file xlisp.fb looks like this:
$magic 0x10130000
$version 2
$name xlisp
$kind procedure
$start
# generated by prof -feedback
# procedure_name file_name freq
xlminit xldmem.c 651846882
xlxgetvalue xlsym.c 564706014
xlabind xleval.c 368782916
xleval xleval.c 360302271
mark xldmem.c 353045832
xlgetvalue xlsym.c 341400298
xlsend xlobj.c 306873567
sweep xldmem.c 232575506
evalhook xleval.c 227803590
gc xldmem.c 216458905
addseg xldmem.c 174118911
evform xleval.c 161070071
xlygetvalue xlsym.c 133714210
xlevlist xleval.c 119441482
xlmakesym xlsym.c 117704318
xldinit xldbug.c 117010681
newvector xldmem.c 113412102
iskeyword xleval.c 105730347 ... |
The procedure_name column indicates the name of the procedure and the file_name column lists the name of the file that contains the procedure. The freq column can be the number of cycles spent in the procedure, the number of times the procedure was executed, or the density (total cycles divided by the procedure size). The cord command places procedures based on the order specified in the feedback file and does not use frequency to determine procedure placement.